MENU :
These 3D homology models were created using www.salilab.org Modeller software.
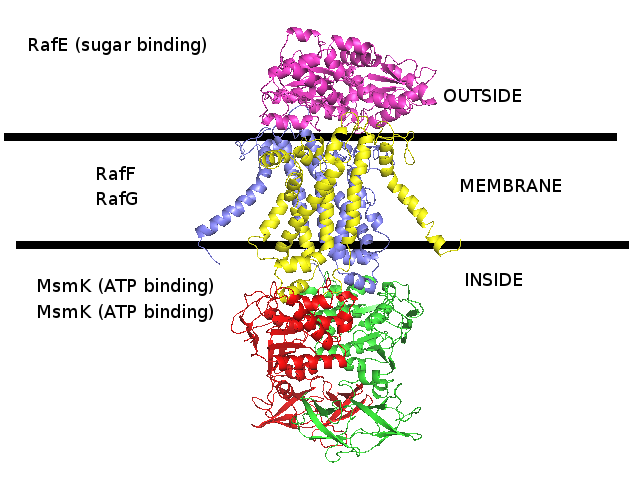
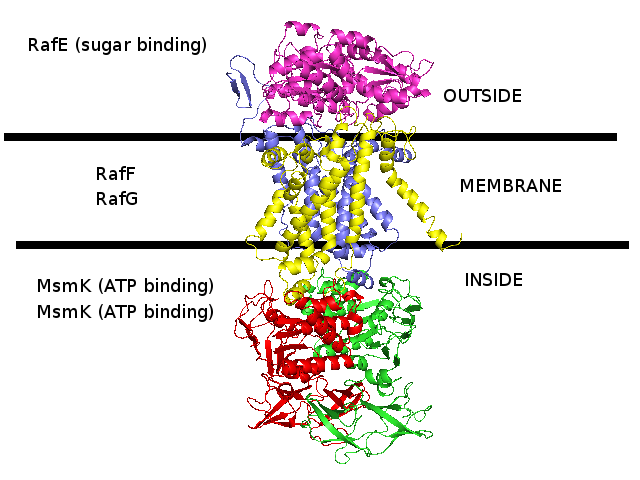
3D homology model MsmRaf2G of Streptococcus pneumoniae TIGR4 MsmK and RafEFG raffinose transporter (TIGR4_MsmRaf2G_2R6G)
- RafE : Entrez Protein NP_346328 : substrate-binding. Modelled on PDBID 2R6G chain E (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- RafF : Entrez Protein NP_346327 : 7 transmembrane helices (TMH). Modelled on PDBID 2R6G chain G (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- RafG : Entrez Protein NP_346326 : 6 transmembrane helices (TMH). Modelled on PDBID 2R6G chain G (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- MsmK : Entrez Protein NP_346026 : ATP-binding cassette. Modelled on PDBID 2R6G chain A (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- MsmK is thought to import raffinose, and RafEFG is also thought to import raffinose, and d-galactose, melibiose, and stachyose.
- There is no experimental evidence that I know of suggesting that MsmK and RafEFG form a transporter complex together.
- Both models MsmRaf and MsmRaf2G are modelled with 2R6G as a template.
The difference between them is that MsmRaf chain F is modelled on 2R6G chain F,
whereas MsmRaf2G chain F is modelled on 2R6G chain G. In both models MsmRaf chain G is modelled on 2R6G chain G.
Energy calculations show that MsmRaf chain F seems to be better modelled by 2R6G chain G and by 2R6G chain F.
PDB file of this 3D homology model
PDB file of this 3D homology model with secondary structure assigned by STRIDE
(with incomplete formatting and information)
PDB file of this 3D homology model with some residues removed
(those residues that protrude from the template were removed for docking purposes)
PDB file of PDBID 2R6G which was used as a template for this 3D homology model
|
|
Periplasmic domain : NP_346328 :
- PF01547 : Bacterial extracellular solute-binding, family 1
- PS01037 : Bacterial extracellular solute-binding family 1, conserved site
-
- G3DSA:3.40.190.10
- PS51257 : PROKAR_LIPOPROTEIN
- SSF53850 : Periplasmic binding protein-like II
- The N-terminal exhibits hydrophobicity as a helix, and is probably a signal peptide,
in which case it should not be shown in the model as it would cleaved off.
|
Membrane domain : NP_346327 :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : ABC_TM1
- 7 transmembrane helices
|
Membrane domain : NP_346326 :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : ABC_TM1
- 6 transmembrane helices
|
Cytoplasmic domain : NP_346026 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- SM00382 : ATPase, AAA+ type, core
- SSF50331 : Molybdate/tungstate binding, transporter activity, ATP binding, MOP-like
- PF08402 : Transport-associated OB, type 2, TOBE_2
- PS00211 : ABC transporter, conserved site
- PD000006 : Q8NZA8_STRP8_Q8NZA8
- G3DSA:2.40.50.100
- G3DSA:3.40.50.300
- PTHR19222:SF42 : SUGAR ABC TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
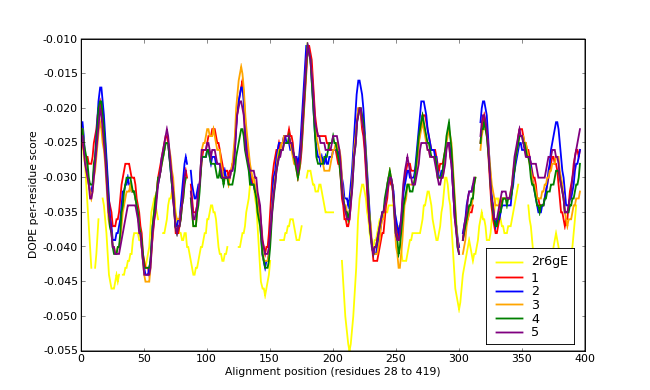
Energy plots of template and homology models from Modeller of NP_346328 (Raf chain E) modelled on 2R6G chain E.
The 1st model (red) was chosen as the final model.
|
|
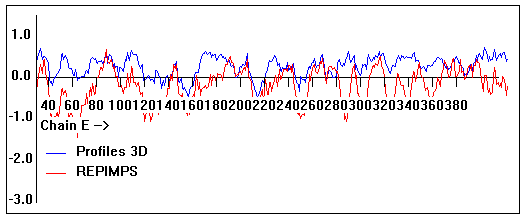
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346328 (Raf chain E) modelled on 2R6G chain E.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
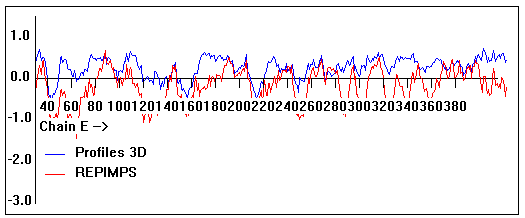
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346328 (Raf chain E) modelled on 2R6G chain E.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
Chain E residues that interface with (are within 3 angstrom of) residues of chain F :
101, 378-382, 384
Chain E residues that interface with (are within 3 angstrom of) residues of chain G :
41-42, 70-72, 190, 254-255, 263, 281
|
|
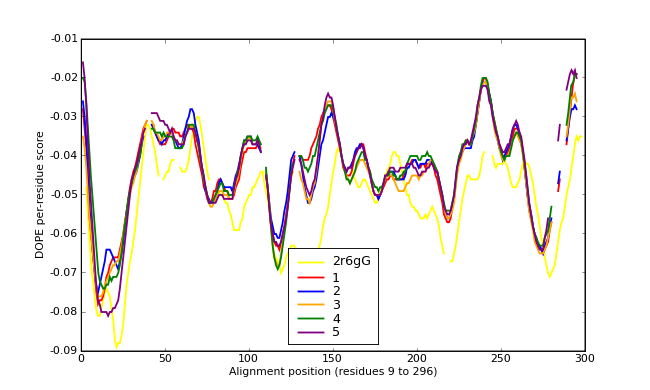
Energy plots of template and homology models from Modeller of NP_346327 (Raf chain F) modelled on 2R6G chain G.
The 1st model (red) was chosen as the final model.
|
|
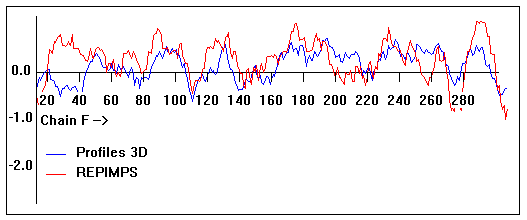
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346327 (Raf chain F) modelled on 2R6G chain G.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
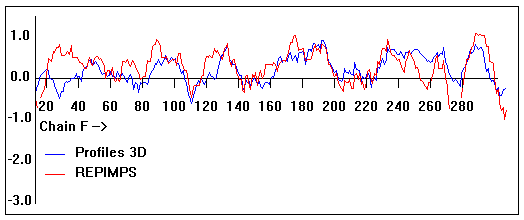
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346327 (Raf chain F) modelled on 2R6G chain G.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
Chain F residues that interface with (are within 3 angstrom of) residues of chain E :
258-263
Chain F residues that interface with (are within 3 angstrom of) residues of chain G :
16, 31, 34-35, 38, 111, 121-134, 176, 179, 183, 186-187, 221, 273, 277, 280, 293,
Chain F residues that interface with (are within 3 angstrom of) residues of chain B :
191-192, 195-201, 205
|
|
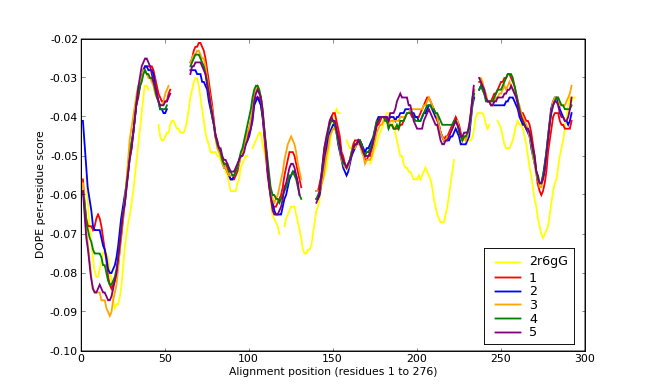
Energy plots of template and homology models from Modeller of NP_346326 (Raf chain G) modelled on 2R6G chain G.
The 1st model (red) was chosen as the final model.
|
|
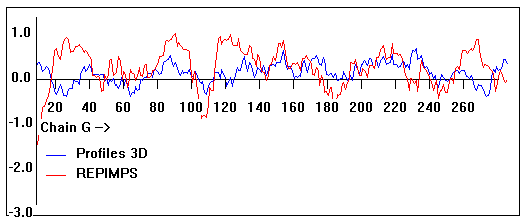
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346326 (Raf chain G) modelled on 2R6G chain G.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
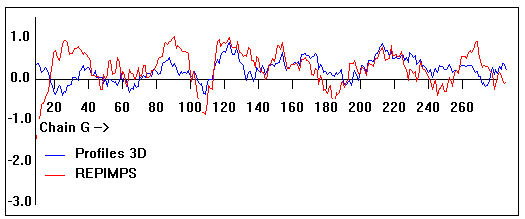
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346326 (Raf chain G) modelled on 2R6G chain G.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
Chain G residues that interface with (are within 3 angstrom of) residues of chain E :
37, 40, 42-43, 214, 224, 231-232, 235-237
Chain G residues that interface with (are within 3 angstrom of) residues of chain F :
29, 40, 100, 112, 114-115, 119-120, 124, 128, 155, 159, 162, 197, 200, 204, 207-208, 230, 244, 247, 248-249, 251-256, 259-260, 262-263, 266-268
Chain G residues that interface with (are within 3 angstrom of) residues of chain A :
166, 170-172, 174-175, 178-179, 185-186, 273-276
|
|
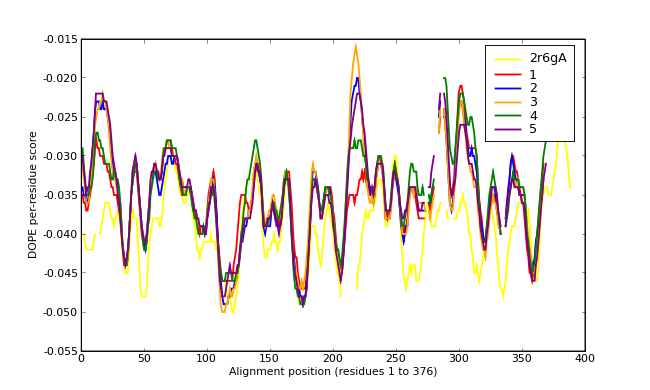
Energy plots of template and homology models from Modeller of NP_346026 (MsmK) modelled on 2R6G chain A.
The 1st model (red) was chosen as the final model.
|
|
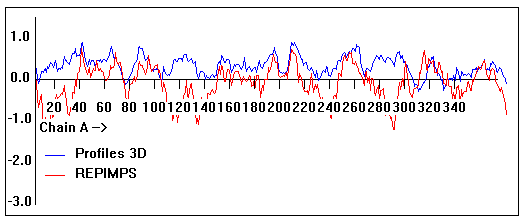
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain A.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
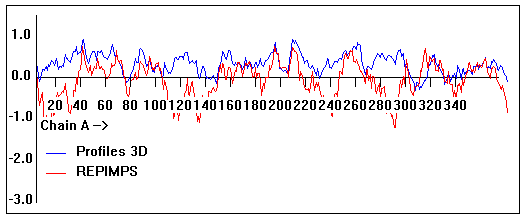
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain A.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
Chain A residues that interface with (are within 3 angstrom of) residues of chain B :
39-40, 143, 161, 164-167, 169, 171, 194, 214, 217, 231, 235, 297, 323-326, 346
Chain A residues that interface with (are within 3 angstrom of) residues of chain G :
49, 54, 75, 83, 85-87, 89, 91-92, 100, 141-142, 152, 165
|
|
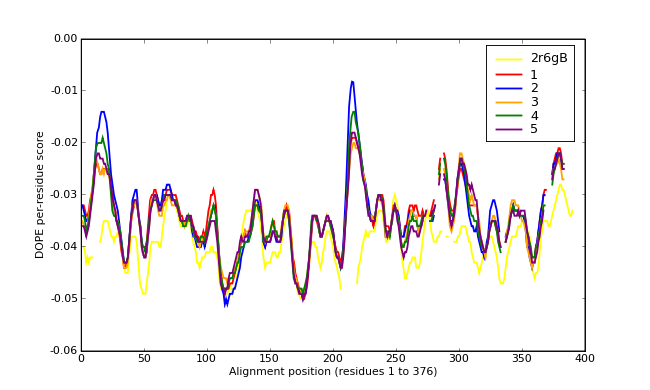
Energy plots of template and homology models from Modeller of NP_346026 (MsmK) modelled on 2R6G chain B.
The 1st model (red) was chosen as the final model.
|
|
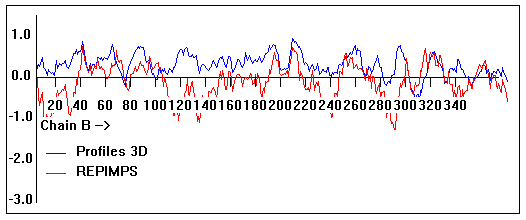
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain B.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
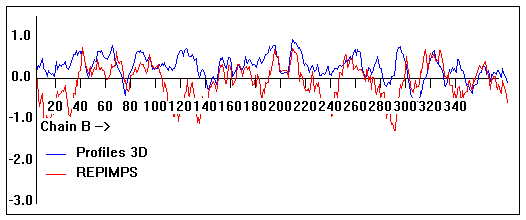
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain B.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
Chain B residues that interface with (are within 3 angstrom of) residues of chain A :
38-40, 127, 143, 161, 165-167, 194, 200, 231, 235, 245, 297-298, 326, 346
Chain B residues that interface with (are within 3 angstrom of) residues of chain F :
52, 54, 74-75, 79, 87-91, 100, 104, 148, 152,
|
|
3D homology model MsmRaf of Streptococcus pneumoniae TIGR4 MsmK and RafEFG raffinose transporter (TIGR4_MsmRaf_2R6G)
- RafE : Entrez Protein NP_346328 : substrate-binding. Modelled on PDBID 2R6G chain E (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- RafF : Entrez Protein NP_346327 : 7 transmembrane helices (TMH). Modelled on PDBID 2R6G chain F (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- RafG : Entrez Protein NP_346326 : 6 transmembrane helices (TMH). Modelled on PDBID 2R6G chain G (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- MsmK : Entrez Protein NP_346026 : ATP-binding cassette. Modelled on PDBID 2R6G chain A (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- MsmK is thought to import raffinose, and RafEFG is also thought to import raffinose, and d-galactose, melibiose, and stachyose.
- There is no experimental evidence that I know of suggesting that MsmK and RafEFG form a transporter complex together.
- Both models MsmRaf and MsmRaf2G are modelled with 2R6G as a template.
The difference between them is that MsmRaf chain F is modelled on 2R6G chain F,
whereas MsmRaf2G chain F is modelled on 2R6G chain G. In both models MsmRaf chain G is modelled on 2R6G chain G.
Energy calculations show that MsmRaf chain F seems to be better modelled by 2R6G chain G and by 2R6G chain F.
PDB file of this 3D homology model
PDB file of this 3D homology model with secondary structure assigned by STRIDE
(with incomplete formatting and information)
PDB file of this 3D homology model with some residues removed
(those residues that protrude from the template were removed for docking purposes)
PDB file of PDBID 2R6G which was used as a template for this 3D homology model
|
|
Periplasmic domain : NP_346328 :
- PF01547 : Bacterial extracellular solute-binding, family 1
- PS01037 : Bacterial extracellular solute-binding family 1, conserved site
-
- G3DSA:3.40.190.10
- PS51257 : PROKAR_LIPOPROTEIN
- SSF53850 : Periplasmic binding protein-like II
- The N-terminal exhibits hydrophobicity as a helix, and is probably a signal peptide,
in which case it should not be shown in the model as it would cleaved off.
|
Membrane domain : NP_346327 :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : ABC_TM1
- 7 transmembrane helices
|
Membrane domain : NP_346326 :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : ABC_TM1
- 6 transmembrane helices
|
Cytoplasmic domain : NP_346026 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- SM00382 : ATPase, AAA+ type, core
- SSF50331 : Molybdate/tungstate binding, transporter activity, ATP binding, MOP-like
- PF08402 : Transport-associated OB, type 2, TOBE_2
- PS00211 : ABC transporter, conserved site
- PD000006 : Q8NZA8_STRP8_Q8NZA8
- G3DSA:2.40.50.100
- G3DSA:3.40.50.300
- PTHR19222:SF42 : SUGAR ABC TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|

Energy plots of template and homology models from Modeller of NP_346328 (Raf chain E) modelled on 2R6G chain E.
The 1st model (red) was chosen as the final model.
|
|
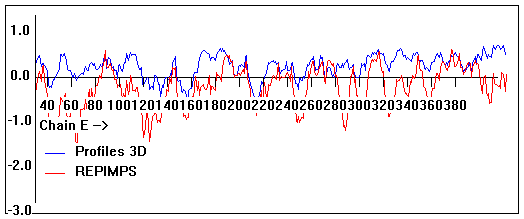
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346328 (Raf chain E) modelled on 2R6G chain E.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
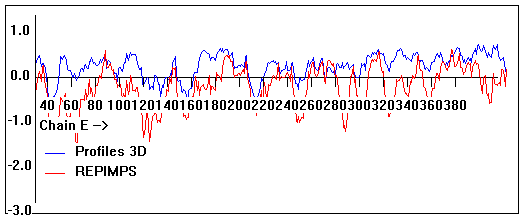
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346328 (Raf chain E) modelled on 2R6G chain E.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
|
|
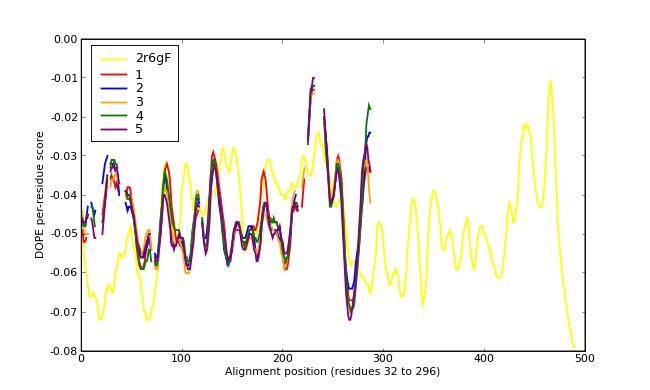
Energy plots of template and homology models from Modeller of NP_346327 (Raf chain F) modelled on 2R6G chain F.
The 1st model (red) was chosen as the final model.
|
|
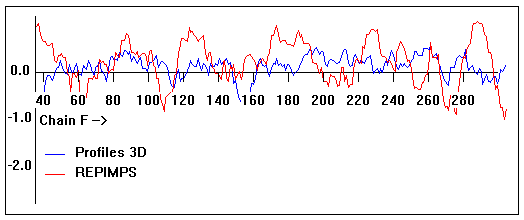
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346327 (Raf chain F) modelled on 2R6G chain F.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
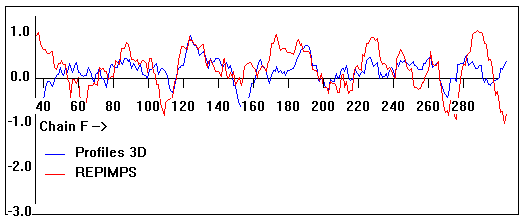
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346327 (Raf chain F) modelled on 2R6G chain F.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
|
|

Energy plots of template and homology models from Modeller of NP_346326 (Raf chain G) modelled on 2R6G chain G.
The 1st model (red) was chosen as the final model.
|
|
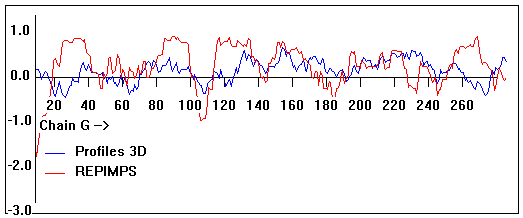
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346326 (Raf chain G) modelled on 2R6G chain G.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
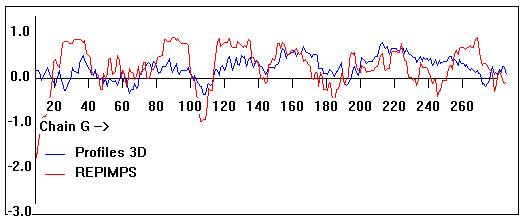
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346326 (Raf chain G) modelled on 2R6G chain G.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
|
|

Energy plots of template and homology models from Modeller of NP_346026 (MsmK) modelled on 2R6G chain A.
The 1st model (red) was chosen as the final model.
|
|
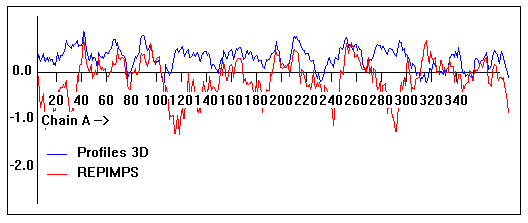
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain A.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
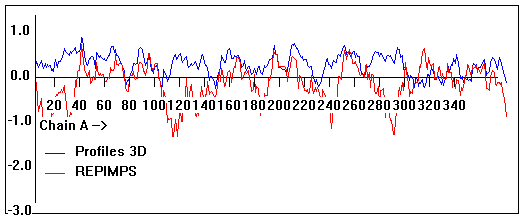
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain A.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
|
|

Energy plots of template and homology models from Modeller of NP_346026 (MsmK) modelled on 2R6G chain B.
The 1st model (red) was chosen as the final model.
|
|
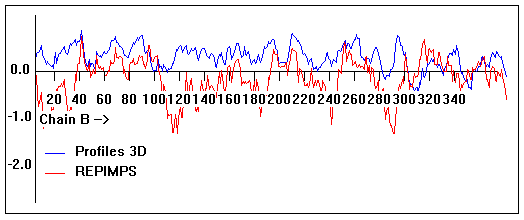
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain B.
Solvent accessible surface area (SASA) calculations done on the chain in isolation, not in the complex with other subunits.
|
|
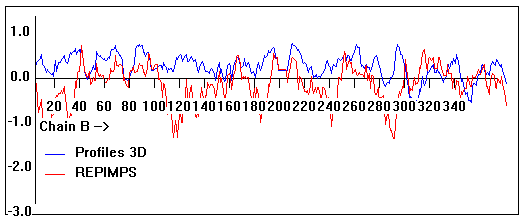
Valpred3D/Profiles3D/REPIMPS plots of final homology model from Modeller of NP_346026 (MsmK) modelled on 2R6G chain B.
Solvent accessible surface area (SASA) calculations done on the chain in the complex with other subunits, not in isolation.
|
|
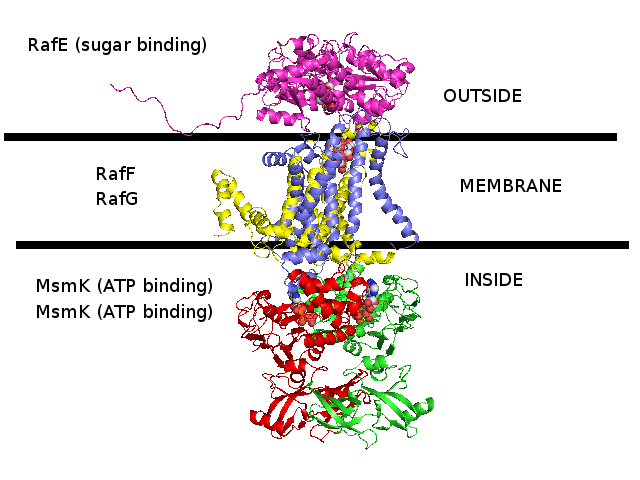
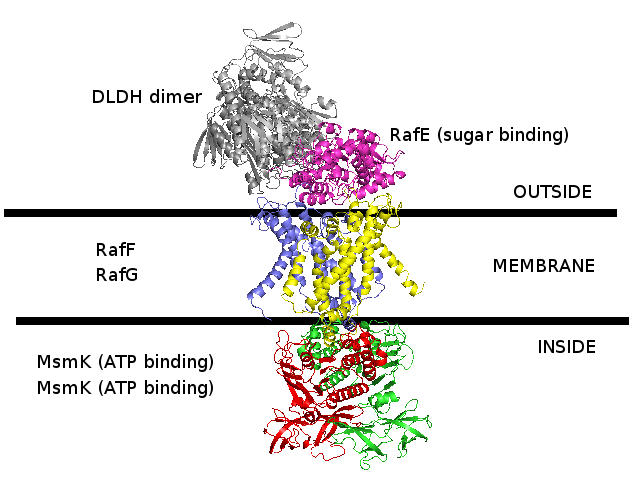
3D homology model 1 of Streptococcus pneumoniae TIGR4 MsmK and RafEFG raffinose transporter (TIGR4_MsmK_RafEFG_2HEU_3FH6_2R6G_2D62)
- RafE : Entrez Protein NP_346328 : substrate-binding. Modelled on PDBID 2HEU chain A (RafE, raffinose transporter, S.pneumoniae).
- RafF : Entrez Protein NP_346327 : 7 transmembrane helices (TMH). Modelled on PDBID 3FH6 chain F (MalF, maltose transporter, E.coli).
- RafG : Entrez Protein NP_346326 : 6 transmembrane helices (TMH). Modelled on PDBID 2R6G chain G (MalG, maltose transporter, E.coli).
- MsmK : Entrez Protein NP_346026 : ATP-binding cassette. Modelled on PDBID 2D62 chain A (multiple sugar binding transport ATP-binding protein, Pyrococcus horikoshii).
- MsmK is thought to import raffinose, and RafEFG is also thought to import raffinose, and d-galactose, melibiose, and stachyose.
- There is no experimental evidence that I know of suggesting that MsmK and RafEFG form a transporter complex together.
This 3D homology model has built them together as a complex due to the following clues :
Here is an operon that might contain the rest of the pieces to the MsmK gene.
MsmK is the ABC-binding part of a raffinose/d-galactose/melibiose/stachyose transporter.
The following operon is for a sugar transporter and lacks an ABC-binding part.
(There is one, but it is for the preceding Ami gene, not the sugar gene here.)
It's sugar-binding protein has an RHS pattern,
and Pyrococcus horikoshii 2D62_A ABC-binding (that is similar in sequence to MsmK)
is on an operon near gene NP_142188 which has the RHS patterns
(just as the Ecoli RHS genes YP_026224/NP_415229/NP_418050 (rhsB/rhsC/rhsR) have these RHS patterns.
In addition, this operon contains "msm operon regulatory protein".
This operon also contains a gene to process alpha-galactosidase - is raffinose an alpha-galactosidase?
Could the nearby AmiD/AmiC oligopeptide ABC transporter be capable of translocating DLDH across the membrane?
- NP_346320 : oligopeptide ABC transporter, permease protein AmiD
- NP_346321 : oligopeptide ABC transporter, permease protein AmiC
- NP_346322 : oligopeptide ABC transporter, oligopeptide-binding protein AmiA
- NP_346323 : hypothetical protein
- NP_346324 : hypothetical protein
- NP_346325 : sucrose phosphorylase
- NP_346326 : sugar ABC transporter, permease protein
- NP_346327 : sugar ABC transporter, permease protein
- NP_346328 : sugar ABC transporter, sugar-binding protein
- NP_346329 : alpha-galactosidase
- NP_346330 : msm operon regulatory protein
PDB file of this 3D homology model
PyMOL file of this 3D homology model
|
|
Periplasmic domain : NP_346328 :
- PF01547 : Bacterial extracellular solute-binding, family 1
- PS01037 : Bacterial extracellular solute-binding family 1, conserved site
-
- G3DSA:3.40.190.10
- PS51257 : PROKAR_LIPOPROTEIN
- SSF53850 : Periplasmic binding protein-like II
- The N-terminal exhibits hydrophobicity as a helix, and is probably a signal peptide,
in which case it should not be shown in the model as it would cleaved off.
|
Membrane domain : NP_346327 :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : ABC_TM1
- 7 transmembrane helices
|
Membrane domain : NP_346326 :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : ABC_TM1
- 6 transmembrane helices
|
Cytoplasmic domain : NP_346026 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- SM00382 : ATPase, AAA+ type, core
- SSF50331 : Molybdate/tungstate binding, transporter activity, ATP binding, MOP-like
- PF08402 : Transport-associated OB, type 2, TOBE_2
- PS00211 : ABC transporter, conserved site
- PD000006 : Q8NZA8_STRP8_Q8NZA8
- G3DSA:2.40.50.100
- G3DSA:3.40.50.300
- PTHR19222:SF42 : SUGAR ABC TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
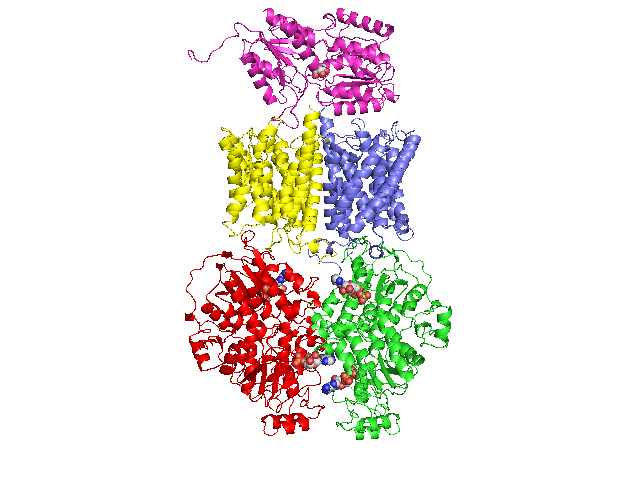
3D homology model 1 of Streptococcus pneumoniae TIGR4 MglA ribose transporter (TIGR4_MglA_2FQX_3B9W_3BK7)
- MglB : Entrez Protein NP_345336 : substrate-binding. Modelled on PDBID 2FQX chain A (PnrA, guanosine-binding, Treponema pallidum).
- MglK : Entrez Protein NP_345339 : 10 transmembrane helices (TMH). Modelled on PDBID 3B9W chain A (Rhesus-homologue, ammonium transporter, Nitrosomonas europaea).
- MglC : Entrez Protein NP_345338 : 9 transmembrane helices (TMH). Modelled on PDBID 3B9W chain A (Rhesus-homologue, ammonium transporter, Nitrosomonas europaea).
- MglA : Entrez Protein NP_345337 : ATP-binding cassette. Modelled on PDBID 3BK7 chain A (ABCE1/RNAase-L Inhibitor protein, Pyrococcus abysii).
- The homologues used to build the 3D homology models for the periplasmic and membrane domains have only weak and unconvincing homology.
The resulting 3D homology models for those domains are therefore probably not good models.
- This S.pneumoniae TIGR4 MglA transporter is thought to transport ribose and other sugars.
PDB file of this 3D homology model
PyMOL file of this 3D homology model
|
|
Periplasmic domain : NP_345336 :
- PF02608 : Basic membrane lipoprotein, lipid binding
- PS00227 : Tubulin conserved site, GTP binding, microtubule
- PS51257 : PROKAR_LIPOPROTEIN
- SSF53822 : Periplasmic binding protein-like I
- 1 signal peptide (not included in model)
|
Membrane domain : NP_345339 :
- PF02653 : Bacterial inner-membrane translocator
- 10 transmembrane helices
|
Membrane domain : NP_345338 :
- PF02653 : Bacterial inner-membrane translocator
- 9 transmembrane helices
|
Cytoplasmic domain : NP_345337 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- PS50893 : ABC_TRANSPORTER_2
- SM00382 : ATPase, AAA+ type, core
- PS00211 : ABC_TRANSPORTER_1
- PD000006 : Q97RG9_STRPN_Q97RG9
- G3DSA:3.40.50.300
- PTHR19222:SF12 : SUGAR ABC TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
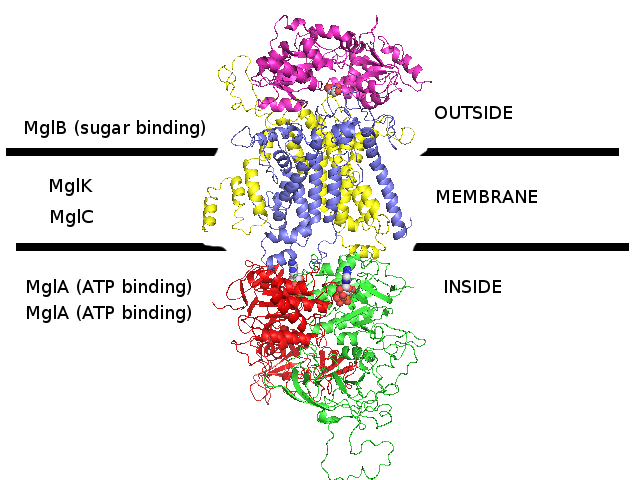
3D homology model 2 of Streptococcus pneumoniae TIGR4 MglA ribose transporter (TIGR4_MglA_2R6G)
- MglB : Entrez Protein NP_345336 : substrate-binding. Modelled on PDBID 2R6G chain E (MalE, maltose-binding, maltose transporter, E.coli).
- MglK : Entrez Protein NP_345339 : 10 transmembrane helices (TMH). Modelled on PDBID 2R6G chain F (MalF, transmembrane, maltose transporter, E.coli).
- MglC : Entrez Protein NP_345338 : 9 transmembrane helices (TMH). Modelled on PDBID 2R6G chain G (MalG, transmembrane, maltose transporter, E.coli).
- MglA : Entrez Protein NP_345337 : ATP-binding cassette. Modelled on PDBID 2R6G chain A (MalA, ATP-binding, maltose transporter, E.coli).
- MglA : Entrez Protein NP_345337 : ATP-binding cassette. Modelled on PDBID 2R6G chain B (MalA, ATP-binding, maltose transporter, E.coli).
- This E.coli maltose transporter that was used as a template for this S.pneumoniae 3D homology model actually does not show any significant sequence homology to the ribose transporter.
It was used as a template because this S.pneumoniae is known to be a sugar transporter and there are no 3D structures in PDB that currently have sequence homology to this S.pneumoniae transporter.
- This S.pneumoniae TIGR4 MglA transporter is thought to transport ribose and other sugars.
PDB file of this 3D homology model
PyMOL file of this 3D homology model
|
|
Periplasmic domain : NP_345336 :
- PF02608 : Basic membrane lipoprotein, lipid binding
- PS00227 : Tubulin conserved site, GTP binding, microtubule
- PS51257 : PROKAR_LIPOPROTEIN
- SSF53822 : Periplasmic binding protein-like I
- 1 signal peptide (not included in model)
|
Membrane domain : NP_345339 :
- PF02653 : Bacterial inner-membrane translocator
- 10 transmembrane helices
|
Membrane domain : NP_345338 :
- PF02653 : Bacterial inner-membrane translocator
- 9 transmembrane helices
|
Cytoplasmic domain : NP_345337 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- PS50893 : ABC_TRANSPORTER_2
- SM00382 : ATPase, AAA+ type, core
- PS00211 : ABC_TRANSPORTER_1
- PD000006 : Q97RG9_STRPN_Q97RG9
- G3DSA:3.40.50.300
- PTHR19222:SF12 : SUGAR ABC TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
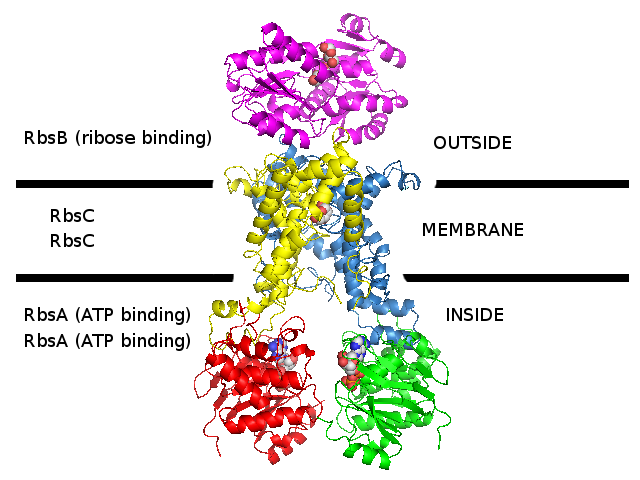
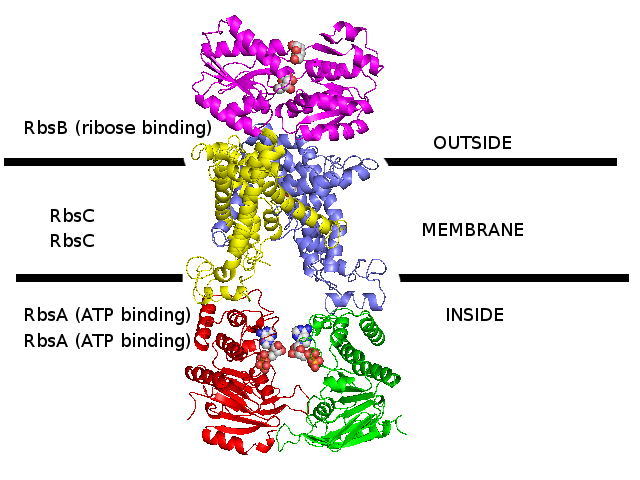
3D homology model 1 of Streptococcus pneumoniae TIGR4 Rbs ribose transporter (TIGR4_Rbs_3LFT_2ONK_1G6H)
- RbsB : Entrez Protein NP_345542 : substrate-binding. Modelled on PDBID 3LFT chain A (RbsB minus N-terminal, S.pneumonia).
N-terminal was modelled on PDBID 2R6G chain F (MalF periplasmic domain, maltose ABC transporter, Escherichia coli).
There is no evidence that N-terminal looks like this.
- RbsC : Entrez Protein NP_345543 : 8 transmembrane helices (TMH). Modelled on PDBID 2ONK chain C (ModB, molybdate ABC transporter, 12 TMH, S.pneumonia).
- RbsA : Entrez Protein NP_345544 : ATP-binding cassette. Modelled on PDBID 1G6H chain A (MJ1267, ABC, Methanococcus jannaschii).
- The complex was assembled according to 2R6G (maltose ABC transporter, E.coli)
- The difference between models 1, 2, and 3 for Streptococcus pneumoniae TIGR4 Rbs ribose transporter is
that different 3D structures were used to model the transmembrane subunit.
The substrate-binding and ATP-binding subunits are the same in all 3 models.
- These S.pneumoniae TIGR4 sequences are the closest sequences that could be found to the E.coli Rbs ribose transporter sequences.
However, these S.pneumoniae TIGR4 sequences are probably not a ribose transporter (perhaps they are an amino acid transporter)
and S.pneumonaie probably does not actually have the Rbs ribose transporter genes.
PDB file of this 3D homology model
PyMOL file (from one merged PDB file)
PyMOL file (from separate subunit PDB files)
PyMOL file (working file used to build and align model subunits)
|
|
Periplasmic domain : NP_345542 :
- PF04392 : ABC transporter substrate binding protein
- SSF53822 : Periplasmic binding protein-like I
- 1 signal peptide (not included in model)
|
Membrane domain : NP_345543 :
- PF02653 : Bacterial inner-membrane translocator
- 8 transmembrane helices
|
Cytoplasmic domain : NP_345544 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- PS50893 : ABC_TRANSPORTER_2
- SM00382 : ATPase, AAA+ type, core
- PS00211 : ABC_TRANSPORTER_1
- PD000006 : Q97QX3_STRPN_Q97QX3
- G3DSA:3.40.50.300
- PTHR19222 : ATP BINDING CASSETE (ABC) TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
3D homology model 2 of Streptococcus pneumoniae TIGR4 Rbs ribose transporter (TIGR4_Rbs_3LFT_2QI9_1G6H)
- RbsB : Entrez Protein NP_345542 : substrate-binding. Modelled on PDBID 3LFT chain A (RbsB minus N-terminal, S.pneumonia).
N-terminal was modelled on PDBID 2R6G chain F (MalF periplasmic domain, maltose ABC transporter, E.coli).
There is no evidence that N-terminal looks like this.
- RbsC : Entrez Protein NP_345543 : 8 transmembrane helices (TMH). Modelled on PDBID 2QI9 chain A (BtuCD, vitamin B12 ABC transporter, 10 TMH, E.coli).
- RbsA : Entrez Protein NP_345544 : ATP-binding cassette. Modelled on PDBID 1G6H chain A (MJ1267, ABC, Methanococcus jannaschii).
- The complex was assembled according to 2QI9 (vitamin B12 ABC transporter, E.coli)
- The difference between models 1, 2, and 3 for Streptococcus pneumoniae TIGR4 Rbs ribose transporter is
that different 3D structures were used to model the transmembrane subunit.
The substrate-binding and ATP-binding subunits are the same in all 3 models.
- These S.pneumoniae TIGR4 sequences are the closest sequences that could be found to the E.coli Rbs ribose transporter sequences.
However, these S.pneumoniae TIGR4 sequences are probably not a ribose transporter (perhaps they are an amino acid transporter)
and S.pneumonaie probably does not actually have the Rbs ribose transporter genes.
PDB file of this 3D homology model
PyMOL file (of the separate subunits)
PyMOL file (working file used to build and align model subunits)
|
|
Periplasmic domain : NP_345542 :
- PF04392 : ABC transporter substrate binding protein
- SSF53822 : Periplasmic binding protein-like I
- 1 signal peptide (not included in model)
|
Membrane domain : NP_345543 :
- PF02653 : Bacterial inner-membrane translocator
- 8 transmembrane helices
|
Cytoplasmic domain : NP_345544 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- PS50893 : ABC_TRANSPORTER_2
- SM00382 : ATPase, AAA+ type, core
- PS00211 : ABC_TRANSPORTER_1
- PD000006 : Q97QX3_STRPN_Q97QX3
- G3DSA:3.40.50.300
- PTHR19222 : ATP BINDING CASSETE (ABC) TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
3D homology model 3 of Streptococcus pneumoniae TIGR4 Rbs ribose transporter (TIGR4_Rbs_3LFT_3D31_1G6H)
- RbsB : Entrez Protein NP_345542 : substrate-binding. Modelled on PDBID 3LFT chain A (RbsB minus N-terminal, S.pneumonia).
N-terminal was modelled on PDBID 2R6G chain F (MalF periplasmic domain, maltose ABC transporter, E.coli).
There is no evidence that N-terminal looks like this.
- RbsC : Entrez Protein NP_345543 : 8 transmembrane helices (TMH). Modelled on PDBID 3D31 chain C (ModBC, molybdate/tungstate ABC transporter, 6 TMH, Methanosarcina acetivorans).
- RbsA : Entrez Protein NP_345544 : ATP-binding cassette. Modelled on PDBID 1G6H chain A (MJ1267, ABC, Methanococcus jannaschii).
- The complex was assembled according to 2R6G (maltose ABC transporter, E.coli)
- The difference between models 1, 2, and 3 for Streptococcus pneumoniae TIGR4 Rbs ribose transporter is
that different 3D structures were used to model the transmembrane subunit.
The substrate-binding and ATP-binding subunits are the same in all 3 models.
- These S.pneumoniae TIGR4 sequences are the closest sequences that could be found to the E.coli Rbs ribose transporter sequences.
However, these S.pneumoniae TIGR4 sequences are probably not a ribose transporter (perhaps they are an amino acid transporter)
and S.pneumonaie probably does not actually have the Rbs ribose transporter genes.
PDB file of this 3D homology model
PyMOL file (of the separate subunits)
PyMOL file (working file used to build and align model subunits)
|

|
Periplasmic domain : NP_345542 :
- PF04392 : ABC transporter substrate binding protein
- SSF53822 : Periplasmic binding protein-like I
- 1 signal peptide (not included in model)
|
Membrane domain : NP_345543 :
- PF02653 : Bacterial inner-membrane translocator
- 8 transmembrane helices
|
Cytoplasmic domain : NP_345544 :
- PF00005 : ABC transporter-like, ATP binding, ATPase activity
- PS50893 : ABC_TRANSPORTER_2
- SM00382 : ATPase, AAA+ type, core
- PS00211 : ABC_TRANSPORTER_1
- PD000006 : Q97QX3_STRPN_Q97QX3
- G3DSA:3.40.50.300
- PTHR19222 : ATP BINDING CASSETE (ABC) TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
The Crystal Structure of the E. coli Maltose Transporter (PDBID 2R6G)
Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J (2007)
Crystal structure of a catalytic intermediate of the maltose transporter.
Nature 450, 515-521. Pubmed 18033289.
|
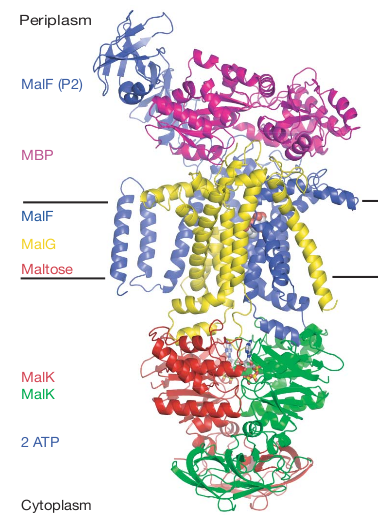
Figure 1. from Oldham et al. 2007, the xray crystal structure for PDBID 2R6G.
|
Periplasmic domain : MBP (maltose binding protein) :
- PF01547 : Bacterial extracellular solute-binding, family 1
- PR00181 : Maltose binding protein
- PS01037 : Bacterial extracellular solute-binding family 1, conserved site
- G3DSA:3.40.190.10
- SSF53850 : Periplasmic binding protein-like II
|
Membrane domain : MalF :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : Binding-protein-dependent transport systems inner membrane component
- 8 transmembrane helices
Membrane domain : MalG :
- PF00528 : Binding-protein-dependent transport systems inner membrane component
- PS50928 : Binding-protein-dependent transport systems inner membrane component
- 6 transmembrane helices
|
Cytoplasmic domain : MalK :
- PF00005 : ABC transporter-like
- PS50893 : ABC_TRANSPORTER_2
- SM00382 : ATPase, AAA+ type, core
- SSF50331 : Molybdate/tungstate binding (MOP-like)
- PF08402 : Transport-associated OB, type 2 (TOBE_2)
- PS51245 : ABC transporter, maltose/maltodextrin import, MalK
- PS00211 : ABC_TRANSPORTER_1
- G3DSA:2.40.50.100
- G3DSA:3.40.50.300
- PTHR19222:SF42 : SUGAR ABC TRANSPORTER
- SSF52540 : P-loop containing nucleoside triphosphate hydrolases
|
|
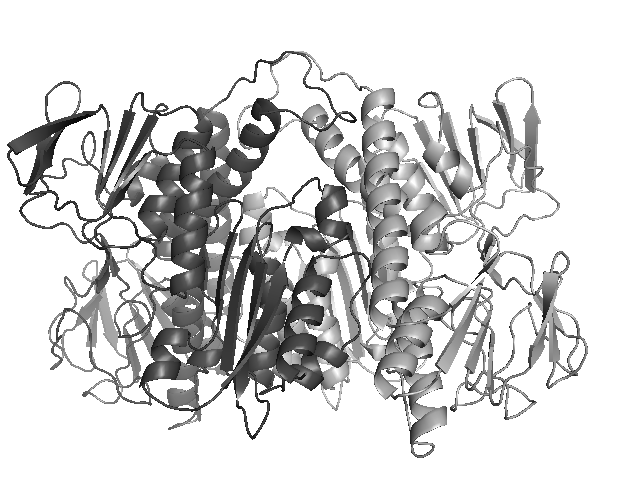
3D homology model Streptococcus pneumoniae TIGR4 DLDH using 2EQ7 as a template (TIGR4_DLDH_2EQ7)
- (not in model) NP_345633 : acetoin dehydrogenase, E1 component, alpha subunit, putative (E1 subunit of pyruvate dehydrogenase complex).
- (not in model) NP_345632 : acetoin dehydrogenase, E1 component, beta subunit, putative (E1 subunit of pyruvate dehydrogenase complex).
- NP_345631 : dihydrolipoamide acetyltransferase. Modelled on PDBID 2EQ7 chain C (E2 subunit of pyruvate dehydrogenase complex).
- NP_345630 : DLDH : acetoin dehydrogenase complex, E3 component, dihydrolipoamide dehydrogenase, putative. Modelled on PDBID 2EQ7 chain A (E3 subunit of pyruvate dehydrogenase complex).
- NP_345630 : DLDH : acetoin dehydrogenase complex, E3 component, dihydrolipoamide dehydrogenase, putative. Modelled on PDBID 2EQ7 chain B (E3 subunit of pyruvate dehydrogenase complex).
- (not in model) NP_345629 : lipoate-protein ligase, putative.
PDBID 2EQ7 was used as a template to make a 3D homology model of S.pneumo DLDH, using the Sali lab Modeller software.
In 2EQ7, DLDH appears as a dimer, and so the 3D homolog model is also a dimer.
RosettaDock was used to dock TIGR4_DLDH_2EQ7 to TIGR4_MsmRaf2G_2R6G.
But first, a manual dock was carried out, to dock DLDH in the general vicinity of the final destination.
The manual dock used the following clues to chose the general vicinity of the final docking destination.
Mande et al. 1996 Structure 4:277-286, PDBID 1EBD says that E2:Pro132,E2:Arg135,E2:Arg139 are conserved
and are involved in starting a binding helix (E2:Pro132) and in electrostatic binding to E3/DLDH (E2:Arg135,E2:Arg139).
Perhaps these conserved E2 residues, that bind DLDH, can be found in the sugar binding subunit of sugar transporters
that are regulated by DLDH and that perhaps also bind DLDH.
In PDBID 2EQ7 chain C, which is the E2 template used for the TIGR_DLDH_2EQ7 model,
these conserved E2 residues correspond to Pro133, Arg137, Lys142.
In TIGR4 raffinose binding protein NP_346328 that is the sequence modelled in the model TIGR4_MsmRaf2G_2R6G,
these conserved residues could correspond to Pro227, Arg230, Lys234,
which conserves a helix initiated by Pro227 which finds itself next to another helix,
with the adjacent helix ends pointing out towards the binding partner and constitute the binding interface.
PDB file of TIGR4 DLDH 3D homology model, manually oriented to bind to the TIGR4 raffinose sugar transporter TIGR4_MsmRaf2G_2R6G.
PDB file of TIGR4 raffinose sugar transporter TIGR4_MsmRaf2G_2R6G, that DLDH has been docked to.
|
|
DLDH dimer
|
|




































